If SETI is all about intelligence, and specifically technology, at the other end of astrobiology is the question of abiogenesis. Does life of any kind in fact occur elsewhere, or does Earth occupy a unique space in the scheme of things? Alex Tolley looks today at one venue where life may evolve, deep inside planetary crusts, with implications that include what we may find “locally” at places like Europa or Titan. In doing so, he takes a deep dive into a new paper from Jeffrey Dick and Everett Shock, while going on to speculate on broader questions forced by life’s emergence. Organisms appearing in the kind of regions we are discussing today would doubtless be undetectable by our telescopes, but with favorable energetics, deep ocean floors may spawn abundant life outside the conventional habitable zone, just as they have done within our own ‘goldilocks’ world.
by Alex Tolley

Are the deep hot ocean vents more suitable for life than previously thought?
In a previous article [1] I explored the possibility that while we think of hot planetary cores, and tidal heating of icy moons, as the driver to maintain liquid water and potentially support chemotrophic life at the crust-ocean interface, radiolysis can also provide the means to do the same and allow life to exist at depth in the crust despite the most hostile of surface conditions. On Earth we have the evidence that there is a lithospheric biosphere that extends to a depth of over 1 kilometer, and the geothermal gradient suggests that extremophiles could live several kilometers down in the crust.
Scientists are actively searching for biosignatures in the crust of Mars, away from the UV, radiation, and toxic conditions on the surface examined by previous landers and rovers. Plans are also being drawn up to look for biosignatures in Jupiter’s icy moon Europa, where hot vents at the bottom of a subsurface ocean could host life. It is hypothesized that Titan may have liquid water at depth below its hydrocarbon surface, and even frozen Pluto may have liquid water deep below its surface of frozen gases. The dwarf planet Ceres also may have a slushy, salty ocean beneath its surface as salts left by cryovolcanism indicate. Conditions conducive to supporting life may be common once we look beyond the surface conditions, and therefore subsurface biospheres might be more common than our terrestrial one.

Image: Rainbow vent field. Credit: Royal Netherlands Institute for Sea Research.
The conditions of heat and ionizing radiation at depth, coupled with the appropriate geology, and water, are energetically favorable to split hydrogen (H2) from water, and then reduce carbon dioxide (CO2) to methane (CH4) via the serpentinization reaction. Chemotrophs feed on this reduced carbon as fuel to power their metabolisms. This reaction has an energy barrier that results in more reactants than products than would be expected at equilibrium. As the reaction energetics are favorable, life also evolves to exploit those reactions, with catalytic metabolic pathways that overcome the energy barrier and allow the equilibrium to be reached, realising the reaction energy..
Biologists now classify life into 3 domains: bacteria, eukaryotes, and the archaea. The bacteria are an extremely diverse group that represent the most species on Earth. They can transfer genes between species, allowing for rapid evolution and adaptation to conditions. [It is this horizontal gene transfer that can create antibiotic resistance in bacteria never previously exposed to these treatments.] The eukaryotes, which include the plants, animals and fungi, range from the single cell organisms such as yeast and photosynthetic cyanobacteria, to complex organisms including all the main animal phyla from spongers to vertebrates. The archaea were only relatively recently (1977) recognized as a distinct domain, separate from the bacteria. Archaea include many of the extremophiles, but perhaps most importantly, exploit the reduction of CO2 with H2 to produce CH4. These archaea are called autotrophic methanogens and require anaerobic conditions. The CH4 is released into the environment, just as plants release oxygen (O2) from photosynthesis. In close proximity to the hot, reducing ocean vent conditions, cold, oxygenated seawater supports aerobic metabolisms, resulting in a biologically rich ecosystem, despite the almost lightless conditions in the abyssal ocean depths.
While CH4 and other reduced carbon compounds are both abiotically and biotically produced, we tend to assume that the formation of biological compounds such as amino acids requires energy that is released from the metabolism of the fixed carbon from autotrophs, whether CH4, or sugars and fats by complex organisms. While this is the case in the temperate conditions at the Earth’s surface, metabolic energy inputs do not appear to be needed under some ocean vent conditions.
The energetics of principally amino acids and protein synthesis is explored in a new paper by collaborators Jeffrey Dick and Everett Shock [2], building on their prior work. The paper examines conditions at two vent fields, Rainbow and Endeavour, compares the energetics of amino acids in those locations, and relates their findings to the proteins of the biota. The two vent fields have very different geologies. The Rainbow vent field is located on the Mid-Atlantic Ridge, at the Azores, and is composed of ultramafic mantle rock that is extruded to drive apart the tectonic plates, slowly widening the Atlantic ocean. In contrast, the Endeavour vent field is located in the eastern Pacific ocean, southwest of Canada’s British Columbia province, and is part of the Juan de Fuca Ridge. It is principally composed of the volcanic mafic rock basalt.
Mafic rocks such as basalt have a silica (SiO2) content of 45-53% with smaller fractions of ferrous oxide, alumina, calcium oxide, and magnesium oxide, while ultramafic peridotites such as olivine have a SiO2 content below 45%, and are mainly comprised of magnesium, ferrous silicate [(Mg, Fe)SiO4]. As a result of the difference in composition and structure, ultramafic rocks produce more hydrogen than the higher SiO2 content mafic rocks.
Typically, the iron sequesters the O2 from the serpentinization reaction to form magnetite (Fe3O4), preventing the H2 and CH4 from being oxidized. The authors use the chemical affinity measure, Ar, to explore the energetic favorability of the production of CH4, amino acids, and proteins. The chemical affinities are positive if the Gibbs free energy releases energy in the reaction, and the reaction is kept further from completing the reaction to equilibrium; that is more reactants and less product than the equilibrium would indicate. Positive chemical affinities indicate that there is energy to be gained from the reaction reaching equilibrium.
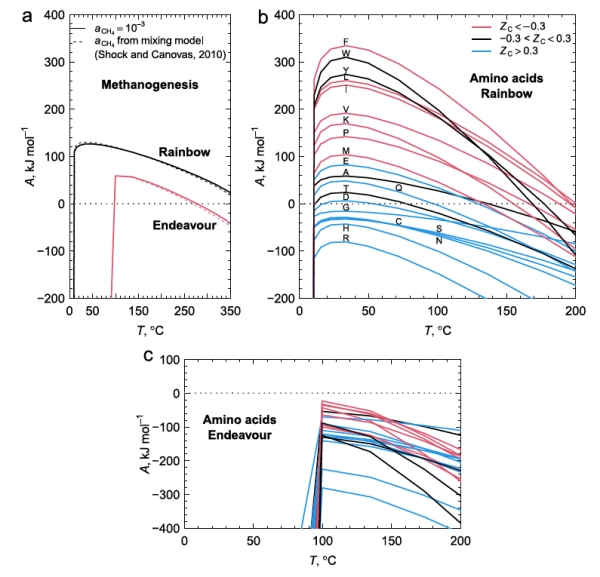
Figure 2 below shows the calculated chemical affinity values for a range of temperatures at the two ocean vent fields Rainbow and Endeavour, at different temperatures as a result of the hot vent water mixing with the cold surrounding seawater. They show that the ultramafic geology at Rainbow has positive affinities for both CH4 and most amino acids, while Endeavour has positive, but lower, affinities for CH4, but negative affinities for amino acids. The Endeavour field not only has lower CH4 affinities for any temperature compared to Rainbow, but this field also has a positive affinity cutoff temperature at about 100C, well above that of Rainbow. As few organisms can live above this temperature, this indicates that methanogens living at Endeavour cannot use the potential free energy of CH4 synthesis to power their metabolisms.
Figure 2b shows that the peak affinities for the amino acids at Rainbow are at around 30-40C, similar to that of CH4. While the range of temperatures where most amino acids have positive affinities at Rainbow to allow organisms to gain from amino acid synthesis, the conditions at Endeavour exclude this possibility in its entirety. As a result, Rainbow vents have conditions that life can exploit to extract energy from amino acid, and hence protein production, whilst this is not available to organisms at Endeavour.
Exploitation of these affinities by life at these two vent fields indicates that autotrophic methanogens will only likely gain metabolic energy from producing CH4 and from anabolic metabolism to produce many amino acids at Rainbow, but not at Endeavour. This would suggest that the Rainbow environment is more conducive to the growth of methanogens, whilst Endeavour offers little competitive advantage against chemotrophs.
Figure 1. The 20 amino acids and their letter codes needed to interpret figure 2b.
Figure 2. a. CH4 production releases more energy at the Rainbow hot vent field with ultramafic geology compared to the mafic Endeavour field when the hot fluids at the event are mixed with cold 2C seawater in greater amounts to reduce the temperature. b. The energetics of amino acid formation at Rainbow. More than half the amino acids are energetically favored. c. All amino acids are not energetically favored at Endeavour primarily due to the much lower molar H2 concentrations at Endeavour.
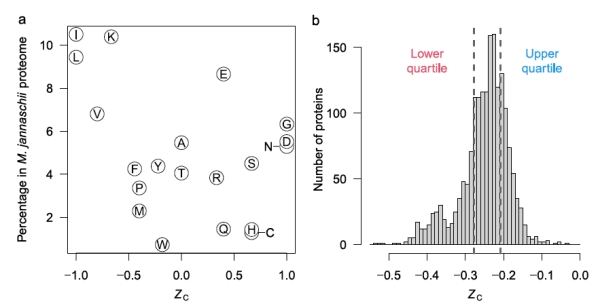
Figure 2b shows that some amino acids release energy when hot 350C water with reactants from Rainbow vents is mixed with cold seawater (approximately 6-10x dilution), while others require energy. The low H2 concentration in samples from Endeavour vents, about 25x more dilute, accounts for the negative affinities across all mixing temperatures at Endeavour. Why might this difference in the affinities between amino acids exist? One explanation is shown in figure 3a, that shows the oxidation values (Zc) of the amino acids. [Zc is a function of the oxidizing elements, charge, and is normalized by the number of carbon atoms of each amino acid. This sets a range of values as [-1.0,1.0].] Notably, those more energetically favored in figure 2b are also those that tend to be least oxidized, that is, they are mostly non-polar, hydrophobic amino acids with C-H bonds dominating.
Figure 3. a. The oxidation level of amino acids. The higher the Zc value, the greater the number of oxidizing and polar atoms composing the amino acid. b. Histogram of all the proteins in the archaean Methanocaldococcus jannaschii based on their per amino acid carbons oxidation score.
Figure 3b shows the distribution of the Zc scores for the proteins of the archaean Methanocaldococcus jannaschii that is found in samples from Rainbow field. The distribution is notably skewed towards the more reduced proteins. The authors imply that this may be associated with the amino acids that have higher affinities and therefore their energy release of formation can be exploited by M. jannaschii.
The paper shows that all the organism’s proteins with their varying amino acid sequences have positive affinities from 0C to nearly 100C. As M. jannaschii has a preferred growth temperature of 85C, its whole protein production produces a net energy gain rather than requiring energy at this vent field, but would not have this energetic advantage if living at Endeavour. While M. jannaschii has an optimum growth temperature of 85C, one might expect other methanogens with optimal growth at lower temperatures closer to those of the optimal affinity values would have a competitive advantage.
As the authors state:
“Keeping in mind that temperature and composition are explicitly linked, these results show that the conditions generated during fluid mixing at ultramafic-hosted submarine hydrothermal systems are highly conducive to the formation of all of the proteins used by M. jannaschii.”
As the archaea already exploit the energetics of methane formation, do they also exploit the favorability of certain amino acids in the composition of their proteins, which are also favored energetically as the peptide bonds are formed?
While figure 3b is an interesting observation for one archaean species found at Rainbow, a natural question to ask is whether the different energetic favorability of certain amino acids is exploited by organisms at the vents by biasing the amino acid sequence of their proteomes, or whether this distribution is common across similar organisms both hot vent-living and surface-living organisms of methanogen archaea and other types of bacteria.
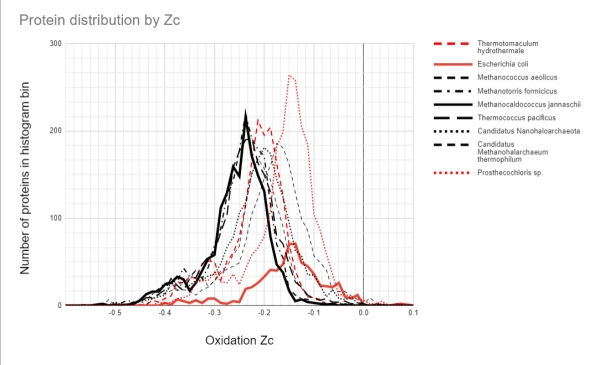
To put the M. jannaschii proteome Zc distribution in context, I have extended the authors’ analysis to other archaea and bacteria, living in hot vents, hypersaline, and constant mild temperature environments. Figure 4 shows the proteome Zc score distribution for 9 organisms. The black distributions are for the archaeans, and the red distributions for bacteria. The distributions for M. jannaschii and the model gut-living bacterium Escherichia coli are bolded.
Figure 4. Histogram of proteome oxidation for various archaea (black) and bacteria (red). Several archaea living in hot temperatures are clustered together. The anaerobic, gut-living E. coli has a very different distribution. The bacterium Prosthecochloris that also lives in the hot vents has a distribution more similar to E. coli, whilst the hot vent living T. hydrothermale has a distribution between the vent-living archaea and E. coli. Two of the archaea also have distributions that deviate from the vent archaeans, one of which is adapted to the hot, hypersaline volcanic pools on the surface. (source author, Alex Tolley)
Figure 4 suggests that the explanation is more complex than simply the energetics as reflected in the proteome’s amino acid composition.
Firstly, the proteome distributions of M. jannaschii and E. coli are very different. They represent different domains of life, inhabit very different environments, and only M. jannaschii is a methanogen. So we have a number of different variables to consider.
Several archaea, all methanogens living in vents at different preferred temperatures, have similar proteome Zc score distributions. The two hypersaline archaea, Canditatus sp., have their distributions biased towards higher Zc scores that may reflect proteomes that are evolved to handle high salt concentrations. One is likely a methanogen, yet its distribution is further biased to a higher Zc score than the other. Of the bacteria, the hot vent-living Thermotomaculum hydrothermale has a Zc score distribution between E. coli and the similar archaean group. It is not a methanogen, but possibly it exploits the amino acid affinities of the hot vent environment.
The other vent-living bacterium, Prosthecochloris sp., has a distribution like that of E. coli. It is a photosynthesizing bacteria similar to green sulfur bacteria, and extracts geothermal light energy. It is not a methanogen. It is found in the sulfur rich smoker vents of the East Pacific Rise.
There seems to be two main possible explanations for the proteomic Zc distributions. Firstly, it may be due to a bias in the selection of amino acids that could release energy when in the H2-rich Rainbow habitat. Secondly, it could be the types of protein secondary structures that are needed for methanogenesis, so that structural reasons are the cause.
Figure 5 shows the protein structures for three approximately matching Zc scores and sequence length for M. jannaschii and E. coli. What stands out is that the lower the Zc score, the more the alpha-helix secondary structure appears in the protein tertiary structure. Both organisms appear to have similar secondary structure compositions when the Zc scores are matched, suggesting that the distribution differences are due to the numbers of proteins with alpha-helix structures rather than some fundamental difference in the sequences. Is this a clue to the underlying distribution?
The amino acids that principally appear in helices are the “MALEK” set, methionine, alanine, leucine, glutamic acid and lysine [6]. A helix made up of equal amounts of each of these amino acids has a Zc score of -0.4, all coincidently in the positive affinity range of amino acids in Rainbow, as shown in figure 2b. This is highly suggestive that the reason for the different distributions is a bias in the production of proteins that have an abundance of alpha-helix secondary structures.
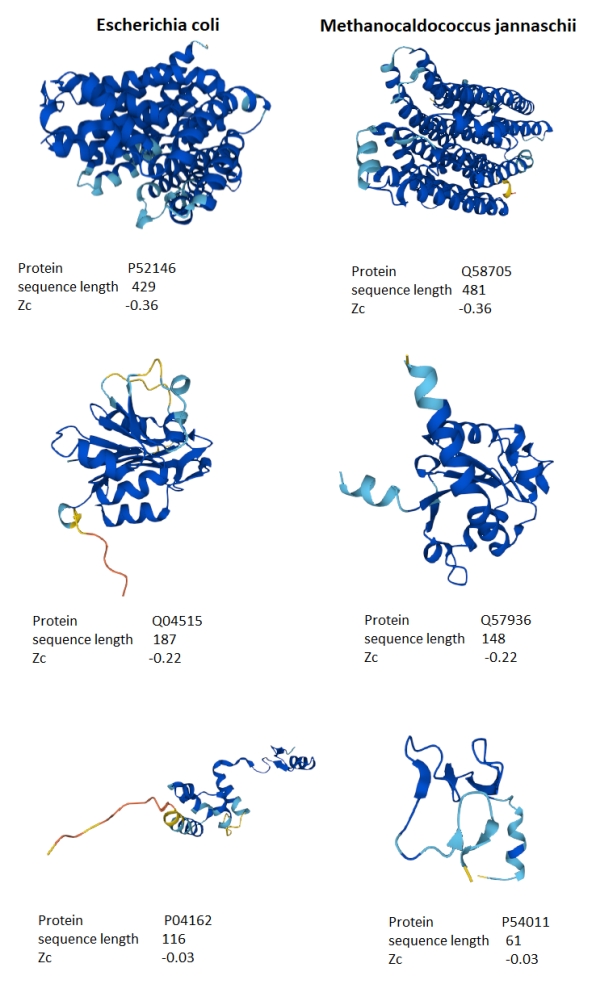
Figure 5. Comparison of selected proteins from M. jannaschii and E. coli spanning the range of Zc scores.
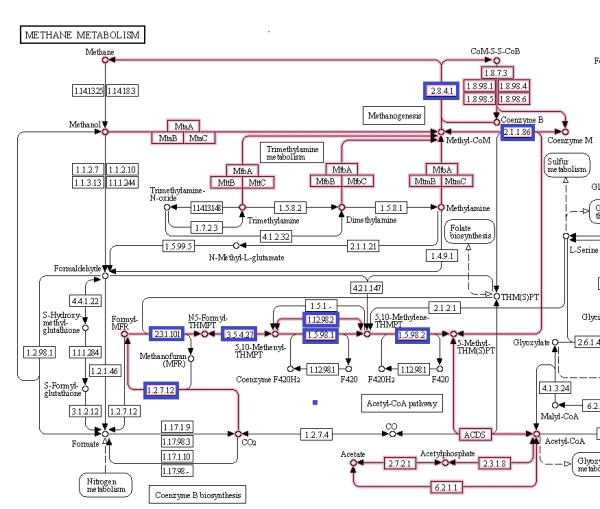
Which proteins might be those with sequences that have higher high alpha-helix structures? As a distinguishing feature of archaea is methanogenesis, a good start is to look at the proteins involved in methane metabolism.
Figure 6 shows the methanogenesis pathways of methane metabolism highlighted. The genes associated with the methanogenesis annotated proteins of M. jannaschii are boxed in blue and are mostly connected with the early CO2 metabolism. From this, some proteins were selected that had tertiary structure available to be viewed in the Uniprot database [3].
Figure 6. Methane metabolism pathway highlighted. Source: Kegg database [4].
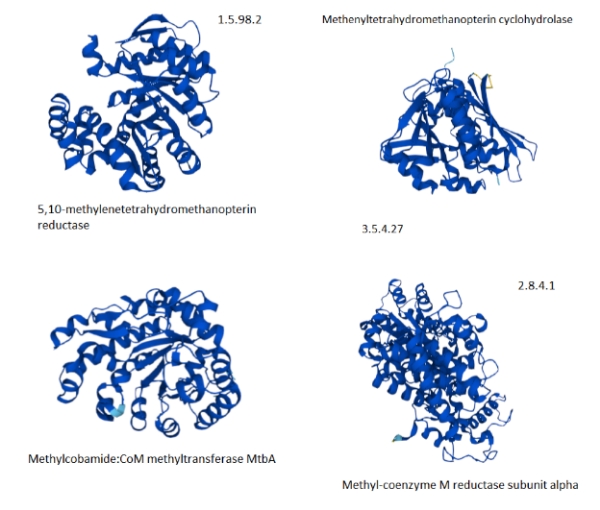
Figure 7. Selected proteins from the methane metabolism pathway of archaea showing the predominance of helix structures [The Kegg #.#.#.# identifiers are shown to map to figure 5.].
The paucity of good, available tertiary protein structures for the methanogenesis pathways makes the selection support a more anecdotal than analytic explanation. The selected proteins do suggest that they are highly composed of alpha-helices. If the methanogenesis pathways are more highly populated with proteins with helical structures, then the explanation of the hot vent-living archaeans might hold.
In other words, it is not, particularly the energetic favorability that determines the proteome composition, but rather the types of metabolic pathways, most likely methanogenesis that is responsible. It should be noted that pathway proteins are not populated by one unique protein as the Kegg pathway indicates where several closely related genes/proteins can be involved in the same function.
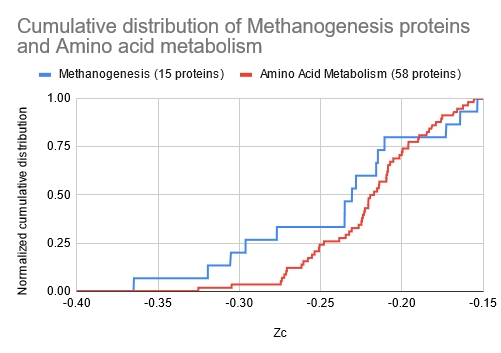
Figure 8. Cumulative distribution of proteins for methanogenesis and amino acid metabolism for M. jannaschii. The methanogenesis proteins are biased towards the lower Zc values, indicating a greater probability of alpha-helix structures.
Figure 7 shows the normalized cumulative distributions of 15 methanogenesis proteins and 58 amino acid metabolizing proteins that have been well identified for M. jannashii.
The distribution clearly shows that there is a bias towards the lower Zc values for the methanogenesis proteins than the more widely distributed amino acid metabolic proteins. While not definitive, it is suggestive that the proteome Zc score distribution between organisms may be accounted for by the presence and numbers of methanogenesis proteins.
Lastly, I want to touch on some speculation on the larger question of abiogenesis. It is unclear whether bacteria or archaea are the older life forms and closer to the last universal common ancestor (LUCA). Because the archaea share some similarities to the eukaryotes, this implies that either the bacteria are the earlier form, or that they are a later form that branched off from the archaea, and the eukaryotes evolved from the archaean branch. The attractiveness of the archaea as the most ancestral forms, as their domain name suggests, is their extremophile nature and their ability to extract energy from the geologic production of H2 to form CH4 as autotrophs, rather than consuming CH4, which has been shown to be relatively out of equilibrium due to the energy barrier to complete the reaction.
If so, does the energetic favorability of amino acid formation at ultramafic hot vent locations suggest a possible route to abiogenesis via a metabolism first model? While the reaction to create amino acids abiotically may be difficult to proceed, they may accumulate over time as long as the reverse reactions to degrade them are largely absent. As peptide bonds are energetically favored, oligopeptides and proteins could form abiotically at the vents as the hot fluids mix with the cold ocean water.
If so, could random small proteins form autocatalytic sets that lead to metabolism and reproduction? A number of experiments indicate that amino acids will spontaneously link together and that they can be autocatalytic for self-replication. Peptides replacing the sugar-phosphate backbone can link nucleobases that also can replicate, the model that was held to be a feature of the RNA World model.
But there is a potential fly in the ointment of this explanation of abiogenic protein formation. The proteins should be formed from amino acids that are composed of both L and D chiral forms. Life has selected one form and is homochiral, a feature that is suggested as a determinant for the origin of any extraterrestrial biologically important molecules detected. Experiments have suggested that any small bias in chirality, due perhaps to the crystal surface structure of the rocks, can lead to an exponential dominance of one chiral form over the other. Ribo et al published a review of this spontaneous mirror symmetry breaking (SMSB) [5].
So we have a possible model of abiotically formed peptides of random amino acid sequences that collect in the pores of rocks at the vents and may be surrounded by lipid membranes. The proteins can both form metabolic pathways and self replicate. If the peptides mostly form self-replicating helices, and these can be co-opted to further extract energy via methanogenesis, then we have a possible model for the emergence of life.
As my earlier article speculated that radiolysis could ensure that chemotrophs in the crust of a wide variety of planets and moons could be supported, we can now speculate that the favorable energetics of amino acid and protein formation may also drive the emergence of life.
As autotrophic organisms like archaea can evolve to exploit the energetics of CH4 and protein production under favorable conditions at seafloor vents, and support the evolving ecosystems of chemotrophs, this suggests that abiotic reactions may have started the process that evolved into the sophisticated methanogenesis pathways of methanogens we see today.
If correct, then life may be common in the galaxy wherever the conditions are right, that is that where ultramafic rocks in the mantle, heated from below by various means, and in contact with cold ocean water exist in combination, whether on a planet similar to the early Earth or possibly at the boundary of the mantle and the deep subsurface oceans of icy moons outside the bounds of the traditional habitable zone.
References
Tolley, A “Radiolytic H2: Powering Subsurface Biospheres” (2021) URL accessed 12/01/2021:
https://www.centauri-dreams.org/2021/07/02/radiolytic-h2-powering-subsurface-biospheres/
Dick, J, Shock, E. “The Release of Energy During Protein Synthesis at Ultramafic-Hosted Submarine Hydrothermal Ecosystems” (2021) JournalJournal of Geophysical Research: Biogeosciences, v126:11.
https://agupubs.onlinelibrary.wiley.com/doi/full/10.1029/2021JG006436
Uniprot database
uniprot.org
Kegg database
genome.jp/kegg/
Ribo, J et al “Spontaneous mirror symmetry breaking and origin of biological homochirality” (2017) Journal of the Royal Society Interface, v14:137
https://royalsocietypublishing.org/doi/10.1098/rsif.2017.0699
Alpha-Helix
https://en.wikipedia.org/wiki/Alpha_helix



Is it possible to compare the biochemistry of submarine vent micro-organisms with those of surface-dwelling analogues to determine which are most likely to have evolved first?
Clearly, smoker metazoans like crustaceans and tube worms may be descended from species that originally evolved in more benign surface environments, but perhaps more primitive single-celled species might contain chemical evidence of an earlier origin. Surely, whether the first living things appeared in the abyss, shallow tidal pools, or in the upper layers of the deep sea must have left some trace molecular evidence.
Perhaps this sort of analysis might give some clues as to where on the early Earth the first living things arose.
One would hope so. (See my response to Robin below for one possibility.)
One attractive feature of hot vents is that they do offer a more stable environment than the surface. No problem of dealing with “snowball” conditions, high surface radiation (e.g. UV), and other environmentally changeable conditions. However, today we see those hot vents associated with plate tectonics. If plate tectonics are key, when did they start? There is no precise timing of this. It could be on either side of the earliest evidence for fossil life.
Another possibility is that the end of the late heavy bombardment indicates the starting point for abiogenesis. Either the conditions became stable enough to allow the first cells to emerge (or to prevent the extinction of earlier life), or subsurface volcanic hotspots around impactors were the first favorable conditions for life to appear.
It is like our biological “dark ages”. I just hope it doesn’t require extensive exploration of the galaxy for young planets to nail down how abiogenesis likely happened on Earth.
I believe this is rooted in the hydrogen hypothesis of endosymbiosis as postulated by William Martin and expanded on by Nick Lane.
Hydrogen production and electron transport may be key in abiogenesis. Electron transport is very much embedded in metabolisms today. It certainly is an interesting hypothesis.
What we need is some “fossil” life that captures this essence without the burden of disentangling it from the evolved metabolisms that we have on Earth today.
The implications of this is that the emergence of the Eukaryote is such a wildly improbably event that it happened only once in the history of our galaxy and possibly the observable universe. This would be the answer to Fermi’s famous question.
Very similar idea to the rare Earth hypothesis that prokaryote life may be common, but complex life is very rare.
Of the explanations for the Fermi Paradox, while biological evolution is one answer, there are so many more.
If true, then the Great Filter is behind us and we have the universe to safely explore with a very low probability of meeting another technological species, possibly any complex life intelligent or not.
Excluding panspermia, the options for the abiogenetic origin of life include deep sea vents and surface waters. Life originating from deep sea vents would have to spread to the surface, acquire mitochondria through ingestion => endosymbiosis => organelle formation with an air-using microbe or equivalent set of cellular molecular machinery.
It may be that the enormous time spans have purged all traces of the deep sea vent specific molecular machinery from modern cells. Yet on a much shorter time scale, malarial parasites carry a few remnant fragments from the photosynthetic machinery of their forebears; a fragment or two of deep sea vent specific machinery in modern cells would indicate that origin.
I would separate the issue of abiogenesis and the acquisition of microbes that become mitochondria. Abiogenesis would lead to a planet of microbes and viruses, including photosynthesizers to escape from the lower energy anaerobic metabolism and to fix the more abundant energy of sunlight.
That the archaea are THE methanogens that exploit the favorable energetics of anaerobic carbon reduction from CO2 to CH4 in the hot vent conditions might be a clue, but clearly, this environment is not exclusive for CH4 production today, making the this metabolism ambiguous for origin of life analysis.
The Dick and Shock paper suggests that amino acid formation is favored in the hot vents. To me, that suggests that the energetics of the amino acid metabolism pathways may be different than in surface living microbes. This might be detectable by inspecting and comparing these pathways in microbes in different environments.
This is a very informative paper, Alex Tolley. I disagree with your conclusions for the following reasons. Quote by Alex Tolley: ” Abiogenesis would lead to a planet of microbes and viruses, including photosynthesizers to escape from the lower energy anaerobic metabolism and to fix the more abundant energy of sunlight.” Isn’t this what happened here on Earth? There is a lot of sunlight on the Earth, which increases the probability.
The one common thing that makes the phylogenic tree of life, bacteria, archaea and prokaryotes possible is the fact that they have DNA and RNA. The tree of life is based on the mutation of the DNA which has 3.1 billion bases pairs. The mutation occurs in the copying process or cell replication is not always exact, but there are errors called point mutations and chromosomal aberrations. One base pair is failed to be copied, so there are two copies, etc. My point is mutation is what allows changes that will make the tree of life able to survive in a new environment. Darwinism only is the empirical, outward, environmental evidence of evolution, but the science of genetics was not known yet at that time. The mutation in the DNA and it’s complexity does explain that all the different forms we see today evolved from the complexity of the DNA through mutation. Another type of mutation is caused by radiation from radioactive isotopes on Earth, and a small amount from the invisible cosmic rays collisions with the molecules of our upper atmosphere which through spallation make debris, the smaller mass relativistic muons raining down from above an 99 percent the speed of light which penetrate tens of meters into the ground. Wiki.
Both DNA and RNA are chemically the same. CHONPS or carbon, hydrogen, OXYGEN, nitrogen, phosphorus, and sulfur. All of the tree of life therefore must need oxygen. The extremophiles and thermophiles have DNA. These still need oxygen. There is oxygen in water, but I think that extremophiles are mutations which could more easily evolve to a more hostile environment, so they migrated there. I don’t think they started there, but are the end product of mutation which migrated there from a less hostile environment.
Extremophiles and thermophiles are from the archaea branch. Humans and animals are from the Eukaryota branch. Bacteria are prokaryotes. These are all different branches of the tree of life as you say, but there differences are the result of mutation, not migration. Also Mitochondria is not an organism or cell itself, but a smaller organelle inside of cell.
I am not clear what you are arguing here. One thing that I think you are assuming is that only DNA/RNA can store the information needed for reproduction and have the structure that allows for mutations, and from that evolution.
We really don’t know that is true. Other polymers may work. The key is encoding, and self replication. I think that experiments that replace the sugar-phosphate backbone indicate that other structures are possible. DNA is certainly a good structure to work with, but it looks good now because it has evolved the various means to translate its information into RNA and thence proteins, and to replicate in various ways. RNA won’t work as a replacement because the strands can form complex structures. What we see with DNA is that it is never separated into single strands except at the point of transcription and duplication. Could the basic structure avoid using oxygen? Possibly by replacing it with sulfur, although sulfur tends to crosslink amino acid chains and might not work. I am not a chemist, so I just don’t know. Could phosphorus replace nitrogen? Possibly, but given its scarcity on Earth, not a good choice compared to nitrogen.
All we do know is that life has evolved from a common ancestor and this version is either the only one, or the last version, or even the most successful that outcompeted all other versions.
One ongoing problem is the evolution of sex. It is not efficient. OTOH, computer simulations show that just mixing alleles and modifying them with crossover during replication can be more powerful as a driver of evolution than mutations. Given that so many organisms engage in sexual reproduction, it seems there must be some benefit over asexual reproduction that is not evident.
As for the order of evolution, whether chemotrophs or autotrophic methanogens came first is unknown, but capturing radiant energy by photosynthesis definitely evolved later. Simply because this source of energy is greater than any other on Earth, photosynthesis has become the primary means of energy capture that supports food chains. This may not be the case on worlds that have less available sunlight for a variety of reasons – from stars dominated by IR light, ionizing radiation forcing life to stay well below the surface, or even worlds where sunlight is unavailable.
I apologize for the mistake. I didn’t know mitochondria evolved from bacteria. I still think there can be no life without abiogenesis.
The idea of taking out the oxygen from the DNA is interesting. I intuitively assume it would be the same thing as damaging some of the base pairs if the oxygen is removed from them. I also don’t know whether or not there could be a working kind of DNA without oxygen.
Infra red radiation is not ionizing because it is long wave. Only short wave radiation is ionizing. Energy is inversely proportional to wavelength, so only the short wavelength radiation is ionizing. It begins in the invisible, ultra violet in the electromagnetic spectrum, so any in the UVC and above, x rays and gamma rays are ionizing meaning that these wavelengths have the power to knock the electron free of an atom which is the same thing as damaging the DNA. For example; A piece of the backbone, the outside of the double helix, or some of the rungs of the ladder in the double helix, the base pairs can be broken with ionizing EMR which can prevent the cell from reproducing.
Maybe chromosomes lead to more complexity and intelligent life?
Infra red is o.k. as long as the exoplanet is in the life belt, but if it is outside and too near the star, then the temperature will be too hot for life and too much infra red will still cook something like those heat lamps.
The backbone of the DNA made of sugar has oxygen. If we remove the oxygen, the backbone of the DNA would be broken. https://foldit.fandom.com/wiki/DNA_backbone
This article says that the backbone of the DNA has Phosphorous bound to oxygen, so if the oxygen is removed, the backbone will be broken. https://foldit.fandom.com/wiki/DNA_backbone
Just as Felisa Wolfe-Simon thought that arsenic could replace phosphorus (she was wrong in that case) I am hypothesizing that sulfur, which is already used as a biological element, might be able to replace oxygen as it has the same outer electron shell as oxygen. Not definitely, but possibly in a different biology elsewhere. Unlike silicon to replace carbon, and arsenic to replace phosphorus, sulfur is already a part of biology both structurally and metabolically (green sulfur bacteria reduce sulfate to H2S just as archaea reduce water to methane.) But I emphasize I am not a chemist and I may be totally wrong about this substitution.
I have papers that show that proteins can substitute for phosphate-sugar coupling of nucleobases, and provide a linear molecule that can self replicate and therefore offer a similar function to DNA.
As for oxygen in the deoxyribose sugar. this illustration shows the difference between ribose and deoxyribose. Note that the bonds with the carbons are not at the carbon with the OH group that differs between the 2 sugars. While DNA has become the information storage molecule primarily in a double-stranded state, and RNA is the means to transfer that information to be translated into proteins, it turns out that single-stranded DNA in some viruses also folds into the same type of folded structures as RNA. They are not that different. Could the OH groups in the sugars be replaced with SH groups instead? IDK, but it is a speculation for life in worlds where sulfur is more dominant than oxygen, or the conditions might favor sulfur rather than oxygen.
Replacing oxygen with sulfur might work for some specialist micro organism like viruses or bacteria trapped in a certain environment since nature works through necessity. The problem is that life did not evolve in sulfur and oxygen is a far more abundant element being found in water H2O and CO2, etc. than sulfur. Life would could not evolve in Earth like conditions without oxygen which is highly reactant being diatomic or composed of two molecules O2, and oxygen having less electrons and less electron shells than sulfur is more reactant than sulfur.
There is also the idea of silicon based life but the oxygen silicon bonds are too strong compared to carbon which is more versatile than silicon. Source. I googled it.
You may well be right that there is no conceivable inhabitable environment where sulfur is more common than oxygen and therefore will not be favored over oxygen in organic molecules.
However, it might be interesting to design such life and place it in an environment where hydrogen sulfide or ammonia replace water. Purely artificial of course, but a thought experiment.
I don’t know if the DNA will still work if we replace all of the oxygen atoms like in the base pairs. Experiments have been made replacing one or two oxygen atoms with sulfur, but not all of them.
Excuse me for the mistake. The oxygen molecule is made of two atoms. O plus O is O2.
The story of eukaryote origins has gotten more interesting – by now I think it is accepted that they are derived from the “Asgard” lineage of eukaryotes. (The use of a capitalized generic name seems confusing to me; I think this originated when they could be detected only as environmental DNA sequences but not cultured in a laboratory) See https://www.frontiersin.org/articles/10.3389/fmicb.2018.01896/full https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6646929/ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7015854/ – the last is the paper where one of them was first cultured after 12 years of attempts. Perhaps coincidentally, these Asgards make their living extracting energy by degrading some amino acids into others. I’m not sure if it’s quite right to say this, but they seem to work with bacteria the way that fungal mycelium works with plants – they have long cellular extensions and work symbiotically with the bacteria. In eukaryotes the bacteria became integral parts of the cell – I wonder if the cellular extensions folded up and merged into a cytoplasm surrounding the nucleus?
Archaea are known as extremophiles for where they live, but bacteria seem to be better at extreme chemistry. Mitochondria and chloroplasts are bacterial in origin. In animals is possible to interface aspects of these chemistries by trading highly oxidized amino acids and related compounds at the mitochondrial membrane. (malate shuttle: https://pathbank.org/view/SMP0000129 )
Looking at overall amino acid composition in terms of oxidation state is very interesting. I do wonder if there are more confounders to account for, especially hydrophobicity. Membrane and wall structure, lipid storage, adhesion to a substrate … there might be something that puts a selective pressure on that. The article does show an oxidation state dependent effect independent of hydrophobicity, but if these components can be teased apart with more data the situation might become clearer.
Not really. Oxygen and other similar elements are very polar, creating a charged surface that interacts with water (e.g. O-H groups). Carbon-hydrogen bonds are non-polar, and hydrophobic, as you would expect from hydrocarbons.
Alpha helices may be composed of more hydrophobic amino acids, but also include hydrophilic AAs at a position to best attract a target molecule in order to hold onto it, or effect some reaction. The Prosite database provides a number of known protein motifs that include those with known functions. Biology really is the best nanotechnology that we know about, but working in ways very different from Drexler’s nanomachines that were once fashionable and the “magic pixie dust” of scifi stories, tv, and movies. There is an industry in directed evolution to increase the power of some of these proteins, e.g. enzymes. No doubt as we get better at computing functionality we will be able to design new proteins de novo including those with novel amino acids and side groups that are extranatural.
Sorry, I should have explained. Look at R versus K and D versus E on Figure 3A. In terms of hydrophobicity there isn’t much of a difference (see https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/midas/hydrophob.html ), but they show a strong trend in agreement with the article. So I think the oxidation state really is a major target of selection here as originally concluded, but if there are also differences in hydrophobicity being selected for at the same time, it could jiggle the numbers and make any trends harder to see.
I see what you are saying. Fair point.
This is an easier table of hydrophobicities to use to demonstrate your point when comparing to the oxidation state of the AAs in figure 3A.
Alex and all, if exolife were discovered, how would a taxonomy be built? How would species be named? I did a quick googlepoke and didn’t find any thought toward this. Would scientist would start with analogous Earth species names? So, for a squid-like form found on Europa:
Europus Cephala Septbrasus Gilsterai?
In the Linnean system the genus name has its first letter capitalized; the species and varietal names are all lower-case.
I suspect no respectable taxonomic authority has weighed in on this. (Taxonomists have something of a reputation for fractious disputes…) More importantly, we just don’t have information. It is entirely conceivable (I’ll leave explanations to the reader) that we could find a sample of Europa water teeming with ciliates and dinoflagellates readily classifiable within an Earthly taxonomy. Or we might find a Domain of life about as distant from bacteria and archaea as they are from one another. Or there could be no recognizable similarity in RNA sequence with anything from Earth. Or they might use PNA as a genetic material. Or they might not actually have species, but activate genetic modules out of a large and varying package of seed material, so that their version of a fish can spawn plants and microbes that work in an ecosystem with it. Or their genes might assemble “on the fly” to make temporary organisms for a brief purpose. Maybe “they” could be vortices in the magnetic field of Jupiter that self-propagate, divide, unite, and send communications that can sometimes turn into magnetic phenomena in other planets. At this point I’ve run out of creativity, but the universe has not. :)
They’ll figure out some naming convention (this antecedent is intentionally left blank). Probably something ugly involving an asterisk or ampersand. &Somethingus otherensis … formally &(Sol::Jupiter::Europa)Somethingus otherensis Guer., assuming you find it first.
Latin and common names are very flexible. They may indicate location, e.g. “australis”, “orientalis”, or form, color, trait, fanciful description, named after a person, etc, etc. or as you indicated. It wouldn’t surprise me if a Europan organism that was identified as part of a terrestrial lineage has “europa” as part of its species naming. If totally novel, then I suspect you will only know it is from Europa by looking up its distribution just as the case is with most terrestrial species today. [Europagraphical distribution map?].
Appreciate this survey article. And it will be well worth referring back to from time to time as these issues of comparing exoplanet environments and terrestrial early life continue to come up. It is perhaps trivial by comparison, but I will note the issue anyway: Since it was so hard for us to locate the Endeavour and other underseavolcanic vent environments, how is it that tube worms and blind crabs find their way to such a place and develop an ecological niche? Their pioneering ancestors were not in their present form, but I don’t think they were archaea either. It seems comparable to tunneling at the quantum scale, or us traveling to the stars on the macroscopic.
Animals blind in dark environments but descended from sighted ancestors usually lost their sight due to the lack of light. And many invertebrates do have microscopic larvae.
The ocean acts as a dispersal mechanism for eggs and larvae, just as the wind will disperse dandelion seeds. Ocean dispersal allows corals to recuperate, and barnacles to attach to ships’ hulls. While most will be eaten by predators or die in unsuitable environments, some will alight at the vents and start a new population there.
R.D.,
Agree here so far. But it still seems a little odd. Previous to the Pacific NW discovery, most fauna examples I had heard of were in cave environments near the surface or on land where fish had made such a transformation ( and of course, correct me if I am mistaken). So in this particular case we have an example off shore at significant (Alvin exploaration type) depth. Were the tube worms and blind crabs pre-existing seabed species further adapted? Or did they originate from farther afield such as from a shoreline genus? At first thought, I did not mean to suggest that blind crabs groped their way to such an environment, but maybe they had after all. On the other hand, crabs with sight originally migrating to such an envrironment seems even more far-fetched. Mote sized crab eggs carried by ocean currents?
It needs some detective work to determine origins. But consider that vent fields may be contiguous with surface islands like Iceland. In such cases, a shallow water organism’s population could migrate to the depths over evolutionary time to improve food sources/avoid predators. Also, don’t forget sea-level changes allow shallow water and land animals to migrate in their contiguous environment which later becomes isolated. This was the case for the marsupial populations of Australia, while the famous Wallace Line in the Pacific is due to a deep water channel that prevented a similar land migration across that line.
Many of the vent organisms would have ancestors that are potentially billions of years in the past, and most species exist for 1-10 million years, allowing evolution lots of time for individuals and sub-populations to become founders in a new environments.
If vents near ultramafic rocks are a better environment for abiogenesis, then maybe an Earth-like planet with a slightly lower silicon fraction might favour the emergence of life. We need not assume that Earth is the most optimal environment for abiogenesis, since there may be worlds out there that are even better.
If the hypothesis for abiogenesis is correct, that is indeed the logical conclusion.
The recent post on White Dwarf Clues to Unusual Planetary Composition suggests that planets with low silica rocks may have existed based on the pollution of WD material after the red giant stage disintegrated the planets. Were these worlds very hospitable to life formation?
I endorse Abelard Lindsey’s recommendation of the work of Nick Lane (and William Martin) in this area. I found Lane’s book “The Vital Question” (https://en.wikipedia.org/wiki/The_Vital_Question) very engaging and quite accessible. If you found the biochemistry here to be rough sledding you might check out this book. In addition, Lane addresses the questions of the evolution of eukaryotes and sex, among many related topics. Highly recommended. The book is from 2015 when the discovery of ultramafic / alkaline hydrothermal vents was (relatively) recent. It is good to see newer work in this important area.
Does Saturn’s moon Mimas have an internal ocean of liquid water?
https://phys.org/news/2022-01-uncovering-evidence-internal-ocean-small.html