With landers on places like Enceladus conceivable in the not distant future, how we might recognize extraterrestrial life if and when we run into it is no small matter. But maybe we can draw conclusions by addressing the complexity of an object, calculating what it would take to produce it. Don Wilkins considers this approach in today’s essay as he lays out the background of Assembly Theory. A retired aerospace engineer with thirty-five years experience in designing, developing, testing, manufacturing and deploying avionics, Don tells me he has been an avid supporter of space flight and exploration all the way back to the days of Project Mercury. Based in St. Louis, where he is an adjunct instructor of electronics at Washington University, Don holds twelve patents and is involved with the university’s efforts at increasing participation in science, technology, engineering, and math. Have a look at how we might deploy AT methods not only in our system but around other stars.
by Don Wilkins
A continuing concern within the astrobiology community is the possibility alien life is detected, then misclassified as built from non-organic processes. Likely harbors for extraterrestrial life — if such life exists — might be so alien, employing chemistries radically different from those used by terrestrial life, as to be unrecognizable by present technologies. No definitive signature unambiguously distinguishes life from inorganic processes. [1]
Two contentious results from the search for life on Mars are examples of this uncertainty. Lack of knowledge of the environments producing the results prevented elimination of abiotic origins for the molecules under evaluation. The Viking Lander’s metabolic experiments provide debatable results as the properties of Martian soil were unknown. An exciting announcement of life detection in the ALH 84001 meteorite is challenged as the ambiguous criteria to make the decision are not quantitative.
Terrestrial living systems employ processes such as photosynthesis, whose outputs are potential biosignatures. While these signals are relatively simple to identify on Earth, the unknown context of these signals in alien environments makes distinguishing between organic and inorganic origins difficult if not impossible.
The central problem arises in an apparent disconnect between physics and biology. In accounting for life, traditional physics provides the laws of nature, and assumes specific outcomes are the result of specific initial conditions. Life, in the standard interpretation, is encoded in the initial period immediately after the Big Bang. Life is, in other words, an emergent property of the Universe.
Assembly theory (AT) offers a possible solution to the ambiguity. AT posits a numerical value, based on the complexity of a molecule, that can be assigned to a chemical, the Assembly Index (AI), Figure 1. This parameter measures the histories of an object, essentially the complexity of the processes which formed the molecule. Assembly pathways are sequences of joining operations, from basic building blocks to final product. In these sequences, sub-units generated within the sequence combine with other basic or compound sub-units later in the sequence, to recursively generate larger structures. [2]
The theory purports to objectively measure an object’s complexity by considering how it was made. The assembly index (AI) is produced by calculating the minimum number of steps needed to make the object from its ingredients. The results showed, for relatively small molecules (mass?<?~250 Daltons), AI is approximately proportional to molecular weight. The relationship with molecular weight is not valid for large molecules greater than 250 Daltons. Note: One Dalton or atomic mass unit is a equal to one twelfth of the mass of a free carbon-12 atom at rest.
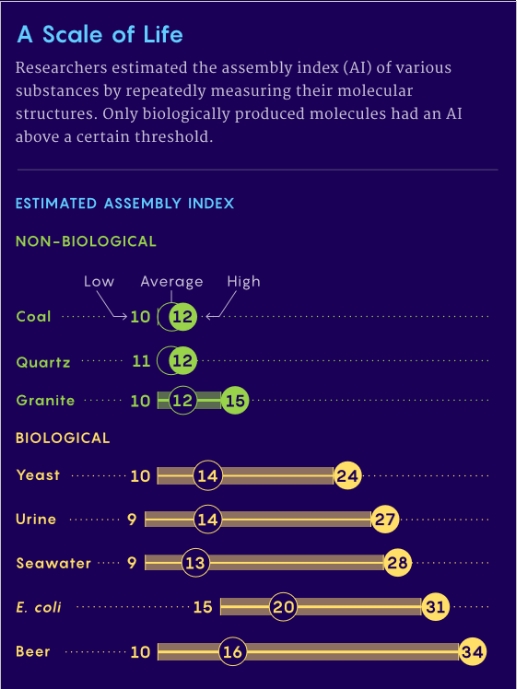
Figure 1. A Comparison of Assembly Indices for Biological and Abiotic Molecules.
Analyzing a molecule begins with basic building blocks, a shared set of objects, Figure 2. AI measures the smallest number of joining operations required to create the object. The assembly process is a random walk on weighted trees where the number of outgoing edges (leaves) grows as a function of the depth of the tree. A probability estimate an object forms by chance requires the production of several million trees and calculating the likelihood of the most likely path through the “forest”. Probability is related to the number of joining operations required or the path length traversed to produce the molecule. As an example, the probability of Taxol forming ranges between 1:1035 to 1:1060 with a path length of 30. In Figure 2, alpha biasing controls how quickly the number of joining operations grows with the depth of the tree.
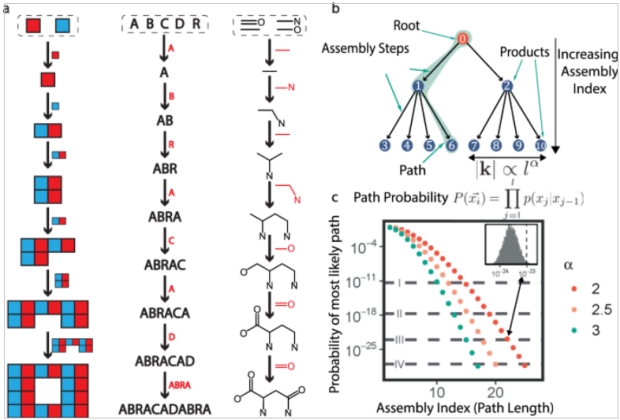
Figure 2. Calculating Complexity
AT does not require extremely fine-tuned initial conditions demanded in the physics-based origins of life. Information to build specific objects accumulates over time. A highly improbable fine-tuned Big Bang is no longer needed. AT takes advantage of concepts borrowed from graph (networks of interlinked nodes) theory. [3] According to Sara Walker of Arizona State University and a lead AT researcher, information “is in the path, not the initial conditions.”
To explain why some objects appear but others do not, AT posits four distinct classifications, Figure 3. All possible basic building block variations are allowed in the Assembly Universe. Physics, temperature or catalysis are examples, constraining the combinations, eliminating constructs which are not physical in the Assembly Possible. Only objects that can be assembled comprise the Assembly Contingent level. Observable objects are grouped in the Assembly Observed.
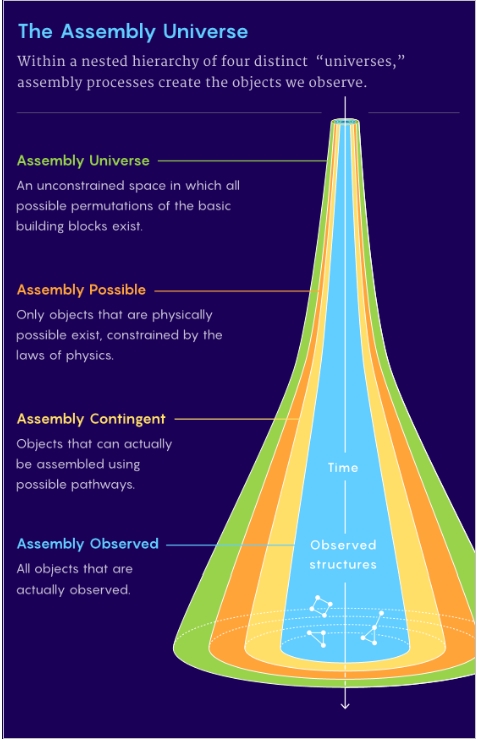
Figure 3. The four “universes” of Assembly Theory
Chiara Marletto, a theoretical physicist at the University of Oxford, with David Deutsch, a physicist also at Oxford, are developing a theory resembling AT, the constructor theory (CT). Mimicking the thermodynamics Carnot cycle, CT uses machines or constructors operating in a cyclic fashion, starting at a original state, processing through a pattern until the process returns to the original state to explain a non-probabilistic Universe.
A team headed by Lee Cronin of the University of Glasgow and Sara Walker proposes AT as a tool to distinguish between molecules produced by terrestrial or extraterrestrial life and those built by abiotic processes. [4] AT analysis is susceptible to false negatives but current work produces no false positives. After completing a series of demonstrations, the researchers believe an AT life detection experiment deployable to extraterrestrial locations is possible.
Researchers believe AI estimates can be made using mass or infrared spectrometry. [5-6] While mass spectrometry requires physical access to samples, Cronin and colleagues showed a combination of AT and infrared spectrometry sensors similar to those on the James Webb Space Telescope could analyze the chemical environment of an exoplanet, possibly detecting alien life.
References
[1] Philip Ball, A New Idea for How to Assemble Life, Quanta, 4 May 2023,
https://www.quantamagazine.org/a-new-theory-for-the-assembly-of-life-in-the-universe-20230504?mc_cid=088ea6be73&mc_eid=34716a7dd8
[2] Abhishek Sharma, Dániel Czégel, Michael Lachmann, Christopher P. Kempes, Sara I. Walker, Leroy Cronin, “Assembly Theory Explains and Quantifies the Emergence of Selection and Evolution,”
https://arxiv.org/abs/2206.02279
[3] Stuart M. Marshall, Douglas G. Moore, Alastair R. G. Murray, Sara I. Walker, and Leroy Cronin, Formalising the Pathways to Life Using Assembly Spaces, Entropy 2022, 24(7), 884, 27 June 2022, https://doi.org/10.3390/e24070884
[4] Yu Liu, Cole Mathis, Micha? Dariusz Bajczyk, Stuart M. Marshall, Liam Wilbraham, Leroy Cronin, “Ring and mapping chemical space with molecular assembly trees,” Science Advances, Vol. 7, No. 39
https://www.science.org/doi/10.1126/sciadv.abj2465
[5] Stuart M. Marshall, Cole Mathis, Emma Carrick, Graham Keenan, Geoffrey J. T. Cooper, Heather Graham, Matthew Craven, Piotr S. Gromski, Douglas G. Moore, Sara I. Walker, Leroy Cronin, “Identifying molecules as biosignatures with assembly theory and mass spectrometry,” Nature Communications volume 12, article number: 3033 (2021)
https://www.nature.com/articles/s41467-021-23258-x
[6] Michael Jirasek, Abhishek Sharma, Jessica R. Bame, Nicola Bell1, Stuart M. Marshall,Cole Mathis, Alasdair Macleod, Geoffrey J. T. Cooper!, Marcel Swart, Rosa Mollfulleda, Leroy Cronin, “Multimodal Techniques for Detecting Alien Life using Assembly Theory and Spectroscopy,” https://arxiv.org/ftp/arxiv/papers/2302/2302.13753.pdf



Coal is biological – originating from coal forests that covered much of the Earth’s tropical land areas during the late Carboniferous and Permian times.
Relative complexity could point to a biological process but not necessarily conclusive.
Perhaps an objective quantitative approach to detecting life signs could be along the lines of a local zone of low-entropy homeostasis? Could that be a more objective definition of life?
I have followed Cronin’s work for a number of years. IMHO, AT is one of the most innovative ways to detect life as it is biologically unbiased compared to terrestrial life biosignatures.
The current method should be designed to work within the constraints of deep space probes to test molecules, e.g. in the plumes on Enceladus, and perhaps a subsurface lake on Mars. If spectroscopy can be done with exoplanet data as suggested, this could be an important tool.
One thing that that was not mentioned in this article ( and others elsewhere) that the relative abundance of the molecule is important. For example complex sugars have very high mol. weights, but their configuration can be quite random. OTOH a chlorophylls have just a relatively few configurations and stand out compared to other possible configurations. It may be that this feature is incorporated into the probability score, but this isn’t clear. ( I really need to read the methods of the relevant papers).
AFAICT this type of biologically unbiased approach has not been tried elsewhere. Even the list of possible biosignature molecules compiled by Walker[?] was based on terrestrial life, e.g. the small phosphine molecule. Large molecules avoid the problem of abiologic small molecules like amino acids that turn up everywhere and can be produced in the lab (Miller-Urey experiment) I’m pleased to see this approach may be gaining traction.
[Spectroscopic identification needs to be far better than when Fred Hoyle claimed evidence of chlorophyll in interstellar molecular clouds. It was a misidentification possibly due to his beliefs about life in space.]
The citation: “Assembly Theory Explains and Quantifies the Emergence of Selection and Evolution” explains that the Assembly Index (AI) contains information about the copy number, i.e. the frequency a particular molecule has. This distinguishes it from large, but random molecules.
Reference #2 modifies my belief in how copy number is important. In this paper, the copy number (or frequency) of a particular structure is more about detection and signal-to-noise ratios. Instead of a random structure based on similar atom and substructure features, the selection of a few molecular structures from the possible space enhances the signal of those structures. The distinction between using assembly steps of structural units and using Kolmogorov complexity as a measure of molecular complexity is made and, I believe, important.
[In one of the Marshall papers, an analogy is used of a few strings having repeats. This is the method that was used to extract short DNA sequences (~6-8 bases) that are transcription enhancers. The same approach can be used to extract any relatively few repeating signals embedded in a noisy, random sequence.]
I am also struggling with the spectroscopy paper. It isn’t clear to me that this will work as advertised. Maybe it is just early days.
The Marshall paper “Formalising the Pathways to Life Using Assembly Spaces” (ref #3) has suggestions for how assembly could be applied to other domains, such as electronic and sonic signals. This has obvious application in SETI, as well as discriminating sounds (vocalizations, stridulations, and songs) from living creatures vs. natural sources.
[If the Venus Life Finder mission does hint that there is life in the Venusian clouds, this assembly theory could be applied to a proposed later mission use of IR spectroscopy and mass spectrometry to further discriminate between living and non-living complex molecules.]
Seager et al created a catalog of small molecules and indicated which may be biosignatures. Toward a List of Molecules….
I had forgotten that I included Cronin’s work in my CD post Detecting Life On Other Worlds (2018)
That link looks very interesting – unfortunately, that paper is all about compiling a list of chemicals – which is not included. That was said to be at http://www.allmolecules.org … apparently moved to http://seagerexoplanets.mit.edu/ASM/index.html … both of which do not respond. Archive.org tried and I think failed (as usual) to scrape useful content. Is there any useful data available?
(It’s described as a list of 14000 chemicals. You could fit that in under a megabyte if you wanted)
Alex: thank you for following up with Sara Seager to find a new link at http://www.allmols.org . Big thanks also to Paul Gilster for relaying your message to me!
It seems like a worthwhile approach.
Does anyone know, off the top of their heads, the most complex molecule found in nature which does NOT result from metabolic, or other, purely biological processes? “Inorganic” complex molecules formed by industrial processes, such as plastics and polymers or long-chain hydrocarbons , do not count.
Just imagine, a planetoid made of styrofoam, or of PVC.
Well, the distribution of potassium ions on mica seems to form a lovely sort of written library text from the Magratheans or their like … if only we could read it! :) https://www.nature.com/articles/s41467-023-35872-y
Phys.org has a story called “Novel crystal compound melts under ultraviolet light.” So imagine a disintigrating probe-then have AI run the clock back.
Here’s a test case I was thinking of, inspired by the previous article on probe swarms. Let’s try the very old-fashioned scenario that the Sun is a sentient being. Hypothesize that the charged particles of the solar wind represent a perfect swarm of probes that it continually sends out to probe and communicate with its environment. (I think it may receive waves of “sound” in response – see https://phys.org/news/2021-10-solar-earth-magnetosphere-stillness-ensues.html ) Is it possible to apply assembly theory to assess whether the complexity of tangled magnetic field lines, anyons, entangled photons, “photonic molecules” and other such oddities beyond my comprehension are in excess of what can be predicted from a non-living system?
Interesting….it seems like the key will be determining that the large molecules exist. Certainly finding them in sample returns from Mars or farther out would be significant , but barring a lucky interstellar visitor, anything more will rely on telescopes and spectroscopy.
I looked through that last paper, but didn’t see anything about what would be required for reliable detection over such long distances.
One’s view of Mars is the litmus test; even with cameras and sensors jammed right into the dirt, people still think that somewhere in a hidden nook, there will be some evidence of life, if not life itself. And this on a planet literally next door to the only known life in the Universe, where even local panspermia was a possibility!
Now, if one thinks that life on Mars is just out of view of cameras centimeters away, what more can be imagined 4 to 100,000 light years from the camera?
In that view, the Universe could be awash with life we can’t yet see as opposed to the lifeless (though magnificent) desolation it appears to be at face value.
The truth of it exists within this range, though we can rule out obvious life in enough places to conclude at least that the Universe isn’t “fine tuned” for life.
This is my first exposure to AT and I haven’t read the references, so I am going to muzzle my initial, very negative and mildly obscene, response to this and just make two points:
1. If they are saying detecting complex molecules suggests life, well, okay.
2. What am I supposed to be learning from Figure 1? That it is easier to make yeast than E. Coli (or beer)? Yes, the average indices for “Non-Biological” molecules/systems(?) are slightly lower than the “Biological” molecules/systems is that it?
Well, they’re probably on to something and I’m just being grumpy.
Source 2 Figure 1 is frustrating. They assemble molecules from “bonds”, which is to say, they add two building blocks that look like ethane and ethene to get propene. But then they make the ester (lower row, second from right) by combining an alcohol and a carboxylic acid in a dehydration synthesis! In most steps they make one bond at a time, but in the final step, they create two bonds simultaneously.
Nonetheless, they do seem to be measuring the complexity within the molecule somehow. When I tried a different approach I did #1 propene (from ethane and ethene), #2 butadiene #3 benzene #4 dimethyl ether (from two methanols) #5 methyl ethyl ether #6 formyl methanoate #7 attach one of #6 to the benzene #8 attach the other.
There is some arbitrariness to the system (HHH and HHHH have equal assembly indices of 1, because you need HH to make either one from H) but it is an integer count after all. I think for any polymer, the assembly index is the AI of the monomer + log2 of the number of subunits. Where the idea shines is that if you have a polymer (RNA) with four possible bases, the complexity isn’t just the four nucleotides + ln 2 of the length – at least, not unless it is made from blocks of repeated single nucleotides. Instead, you have to figure out how to make each unique sequence contained in the RNA.
I do think though that the mica cleavage planes I mentioned above would take a large number of assembly operations to construct, conceivably approaching those in a piece of RNA. The same should be true of a randomized oligonucleotide construct from the sort of pool you might begin with in an aptamer selection experiment.
Re: beer. Is it a coincidence that the highest estimate of the AI of yeast (24) added to that of beer (34) minus the average AI of beer equals 42? I think not!
Interpreting figure 1.
As coal (and oil) is formed from dead plants and animals, the low score is a false negative, although it nicely distinguishes between extant and extinct life. [If we fund coal or oil on Mars, it would likely indicate that Mars was once living.]
Beer having a higher maximum score than yeast and E. coli. I think this may be due to fewer complex molecular structures in beer and therefore increasing the signal-to-noise ratio, compared to the greater diversity of structures in the 2 organisms.
In the Nature Communications paper, there is a larger set of comparisons. Yeast heated to 200 and 400C have scores similar to coal, i.e. the yeast are now dead, rather than living. As expected, distilled single-malt scotch has a lower score than beer.
It is a pity that various plants were not tested. Chlorophyll would be an obvious molecule to detect. [Would the maximum score decrease for deciduous tree leaves in the Fall?]
What about the chitinous exoskeletons of insects and crustacea?
As with other analytical techniques. there needs to be a catalog for many more biological, and non-biological samples. The apparent clear-cut separation based on the limited sample set may prove not as unambiguous as indicated.
Reading the AT papers again, I realized that the idea was based on string rewriting systems (c.f. The Algorithmic Beauty of Plants. [Cellular automata are the simplest use of string rewriting, e.g. Wolfram’s rules, Conway’s “Game of Life”]. String rewriting, e.g. L-systems have rules that have order precedence, e.g. a rule for a longer string detected is invoked before testing a rule for shorter strings.
AT determines the rules from the data, looking for the smallest repeating molecules and using those units to build larger assemblies.
This makes sense – up to a point. For example, tetrapyrrole rings composed of 4 pyrroles are the core structures of chlorophyll and blood.
Where I have difficulty with the concept is with DNA and proteins. The basic units are nucleobases and amino acids. Any repeating structures are not assembled as units, but incrementally. Therefore I would class both DNA and proteins as having assembly size based on the number of these base units. Thus the urine example must include proteins that leaked from incomplete kidney function. A perfectly efficient kidney would only have urea and salts and these would have a very low assembly index value.
If one is agnostic about how the specific biology operates, then it is acceptable to argue that molecular entities could be assembled from larger repeating smaller assemblies. But then how to determine whether the detected proteins or DNA are not a small number of samples taken from the possible space of sequences? Electrophoresis of proteins will separate proteins by molecular weight and change. That this technique results in fairly distinct bands (1-D) or blobs (2-D) does indicate that the actual proteins must have copy numbers which can be the case if sequences were random. However, with ever smaller samples, at what point are measurements taken of single protein chains that could be single examples from a random sequence space? The greater the possible range of sequences, e.g. of DNA in seawater, the lower the signal-to-noise ratio. That we can isolate DNA fragments and determine species from water samples indicates that our techniques do work, although copy number of DNA is increased by polymerization enzymes, which should maintain the relative ratios of abundances.
Many years ago, it was shown that Zempel-Liv-Welch compression (used in Zip compression) was quite good at discriminating between different sequences and images. This crude method seems quite similar to AT in principle. I would like to see how such an algorithm compares to AT in separating living from dead organisms, as well as abiotic compound mixtures.
There is postulated a continuum from physics to life.
OT. – Although hopefully relevant to SETI.
I was pondering the issue of economic growth and planetary heating. We currently consume about 1 millionth of the energy that reaches the Earth’s surface (but that excludes the use of the sun to grow food and maintain the biosphere).
If we assume that this insolation is the maximum we can use, under varying growth rates either purely exponential or logistic, at our last 70 years of energy growth (5.7% pa) it would take less than 300 years to reach the maximum energy use (K I civ level). Reducing energy growth to 1% pa, we would reach maximum energy use in about 1500 years. Not very long.
That doesn’t bode well for any civilization that needs to grow. Any further growth would require the economy to develop off-planet. However, this only buys us a few millennia before we reach K II status. Growth must slow to a crawl to reach K III.
[For other reasons, we need growth now to avoid stagnation and slipping into a dystopia with rather nasty social systems.]
So here is where I would like some help with the math.
We know that Earth without a heat-trapping atmosphere would have a surface temperature below 0C. This suggests that if the Earth was covered in a shell of solar collectors, the surface of the shell should stay temperate. Inside the sphere, the energy would be used to maintain the biosphere and the human economy. Think of the inside having projections and lighting that mimic the open sky.
Because there is still CO2 inside the shell, it would still get very hot, so the climate would need “air conditioning”. My question is whether large radiators on the outside of the shell could dissipate the excess heat to maintain the temperature inside the shell. I assume the radiators would be equatorial, lying in the plane of the orbit with the surfaces in the north-south orientation, rather like Saturn’s rings. In principle this is an Earth-sized O’Neill colony but with the interior illumination created by artificial lighting rather than with mirrors reflecting sunlight.
[I appreciate that the biosphere would not look natural and many phenomena might have to be artificially created. I also assume that O’Neills cannot be made to maintain a biosphere for long periods like the Earth and that therefore they will of necessity require period biosphere maintenance from the home world, restricting their number and potential for population growth. Why build in space when you have a planet?].
[A rather silly Dan Dare story was about a visit to such a planet (“The Platinum Planet”),]
SETI implications.
1. Civilizations would be constrained to the KI level with zero energy growth. [Although this does not imply an absence of change in the culture and its technology.]
2. Remote viewing by telescope would show no living biosphere on the planet, but the spectra would indicate the planet is a megastructure – just a lot smaller than a Dyson sphere/swarm, but much larger than our conceptions of detectable megastructures.
3. Depending on the orientation and absorption efficiency, would we see the planet as an IR emitter, but only if there was some tilt of the plane to our system? Seen edge-on, the planet would be invisible except in transit.
4. Speculating, does this extend a civilization’s lifetime at a KI level, especially as it has greater climate control possibilities over deep time and could stave off the slow luminosity increase of its star.